Single Molecules, Single Strands
Thu Dec 18, 2014
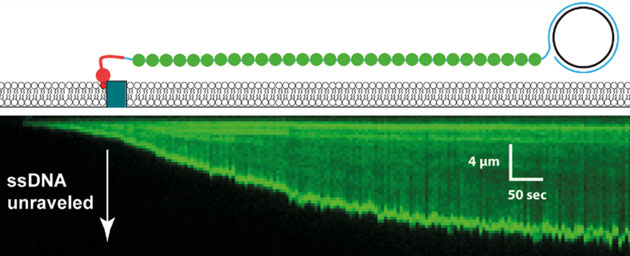
Unravelling of a single-stranded DNA generated by PCR and visualized by a fluorescent single stranded DNA-binding protein.
Today’s Chemical Biology Lecture was by Eric Greene who spoke of his work on using “DNA curtains” to investigate the DNA search mechanism of the RAD51 dna-repair enzyme. The basic idea is that when the cell notices a double strand, it uses the other chromosome’s copy of the same region to fill the gap in. The process is complicated but the first step is to find the other copy and successfully align the two strands.
Although we all know that base-pairing is great and works (ahem…PCR, cloning), the searching mechanism is still rather mysterious: how do the repair enyzmes sample the billions of basepairs needed to find the copy to use for repair? Its a needle-in the haystack problem and Eric’s contribution to understanding it was a delight to hear. Since I saw him speak a few years ago it seems his technology has really matured and it allow him to approach the question in a unique, and visually stunning way.
Eric’s lab studies DNA-binding proteins using a technology he has developed that is truly one-of-a-kind. The basic idea is to use internal reflection microscopy to study DNA strands that are tethered to the top surface of a flow cell, the contents of which can be modified. He was able to use rolling displacement amplification of a target sequence to generate long single stranded DNA, which was coated with a ssDNA-binding proteins and affixed to the inside of the flowcell chamber. These ssDNA-binding proteins were displaced upon the addition of RAD51 and ATP. To this flow cell he can now add labelled dsDNA oligonucleotides which RAD51 will use to search for homologous sequences which can be visualized by the appearance of the fluorescent oligos. The DNA curtains allow Eric and his team to to watch thousands of events simultaneously and track the binding over time. He showed us convincing data that eight base-pairs are needed for the initial binding: the average occupancy time between less-than eight and eight was somewhere between three and four orders of magnitude! What! That some binding-energy black-magic right there and it represents a very important and yet-unknown structural change to stabilize the complex. The significance of that is also to allow a rapid sampling where 8bp similarity is required, effectively eliminating non-sepcifi binding to >90% of chromosomal DNA. After the 8bps, the addition of extra complementary nucleotides led to increased binding in a stepwise fashion that was consistent with the Paveletich structure of ssDNA-RAD51 complex showing an extended (non B-from DNA) ssDNA where normal base stacking is interrupted except where the basepairs are stacked in groups of three. This suggests energy landscapes that jump in groups of three base-pairs after the initial 8bp requirement had been met.
The upshot of all of the data Eric presented was a theory about “microhomology” searching where short bits of homology drive the search process for homologous sequences. This was a beautiful piece of work that elucidates aspects of the basic biophysics of DNA-binding proteins and suggests that the DNA-curtain approach may be maturing to the point of making very specific predictions about the actions of DNA binding proteins.
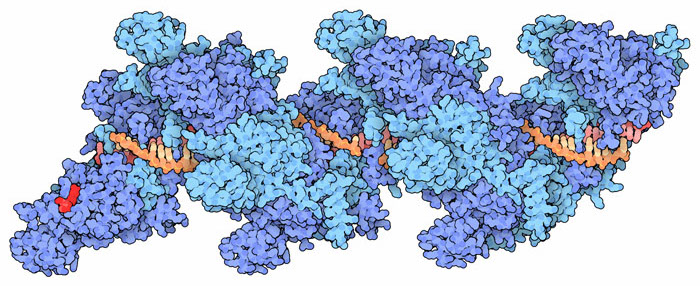
RAD51 and ssDNA structure courtesy of David Goodsell and the PDB
