Visualizing Mibig Gene Clusters with Graphviz
Wed Aug 17, 2016
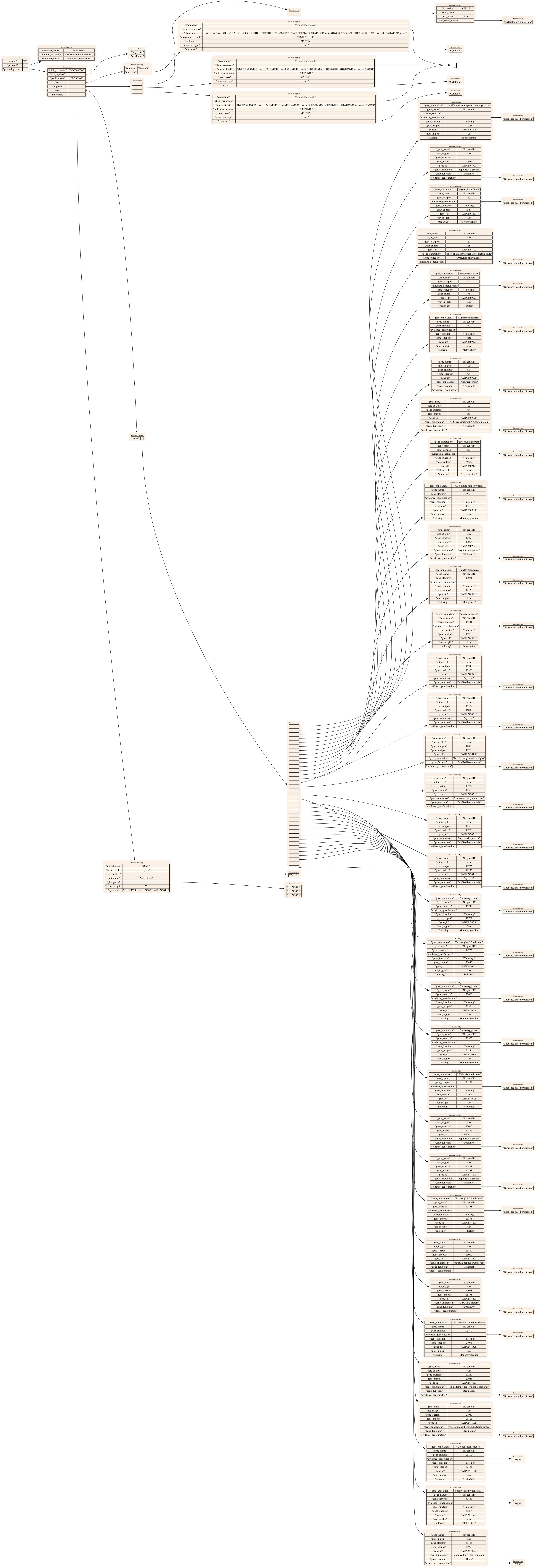
Mibig is an awesome resource for the natural product community by providing a repository and a standard for information about biosynthesis. The only complaint I have is that I think the schema is a bit complicated. This is not a knock on the designers: its a lot of information that has to be wired up somehow or another and they’ve done an excellent job by successfully putting it together. But as it gets used a bit more I think we may be able to improve it. To get a better handle on the Mibig metadata, I’ve downloaded the JSON and visualized it with Graphviz using a clojure-based toolchain: datascope->rhizome->graphviz). The data/images are uploaded to a repo where you can view the graphs.
I’ve chosen a few examples to post here to give an idea about how the data is structured - big NRPS/PKS clusters with lots of genes and with module information as well as numerous small or incompletely annotated clusters. Down the line I would love to tidy this data into triplets and incorporate into a datomic/datascript DB for querying using datalog. I think that would be immensely powerful tool but it would require simplification of the data structures. As a first pass attempt to get the nested structures into a datalog-friendly format I also tried Alan Dipert’s intension library which looked promising but is really geared for relatively shallow nested datastructures. To truly incorporate the Mibig data into datalog we would ideally spec the input JSON/EDN, write or conceive of a datomic/datascript schema and then write some custom translation functions to ingest the data. Its something I am curious to try but represents a reasonable time investment. In order to maximize the power of datalog, I imagine I would try to flatten the data as far as possible by trying to squeeze down the number of entities - clusters, organisms, compounds, genes, modules - and trying to define all possible attributes on those types in terms of keys. I think the intrinsic power of datalog to handle ragged data would allow you to specify all cluster-type-specific as attributes on the cluster level and then get some really informative querying across the dataset. But for now some pics.
Note: Browsing the pics on github will let you zoom in for more detail.
BGC 200: Ariemitemycin (Type II Polyketide)
BGC 685: Brassiscicene (Terpene)
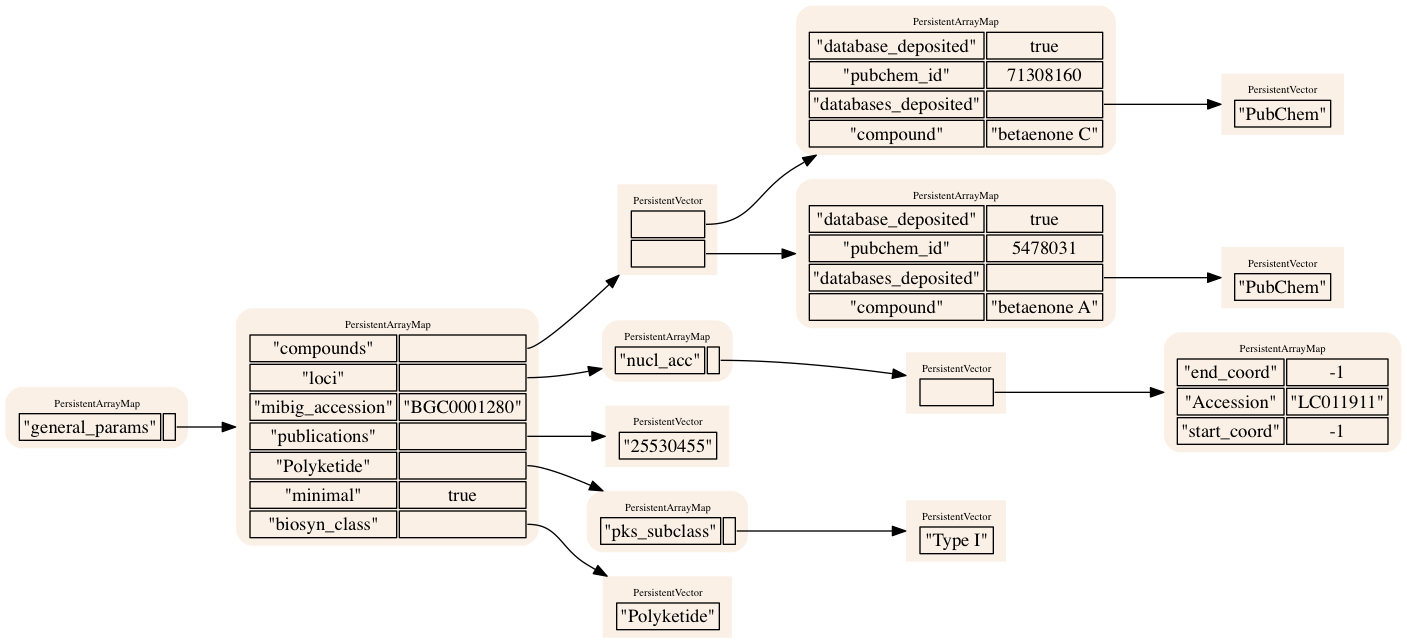
BGC 1064: Cyclindrocyclophane (Polyketide)
BGC 1280: Betaenone A

