structural biology and natural products
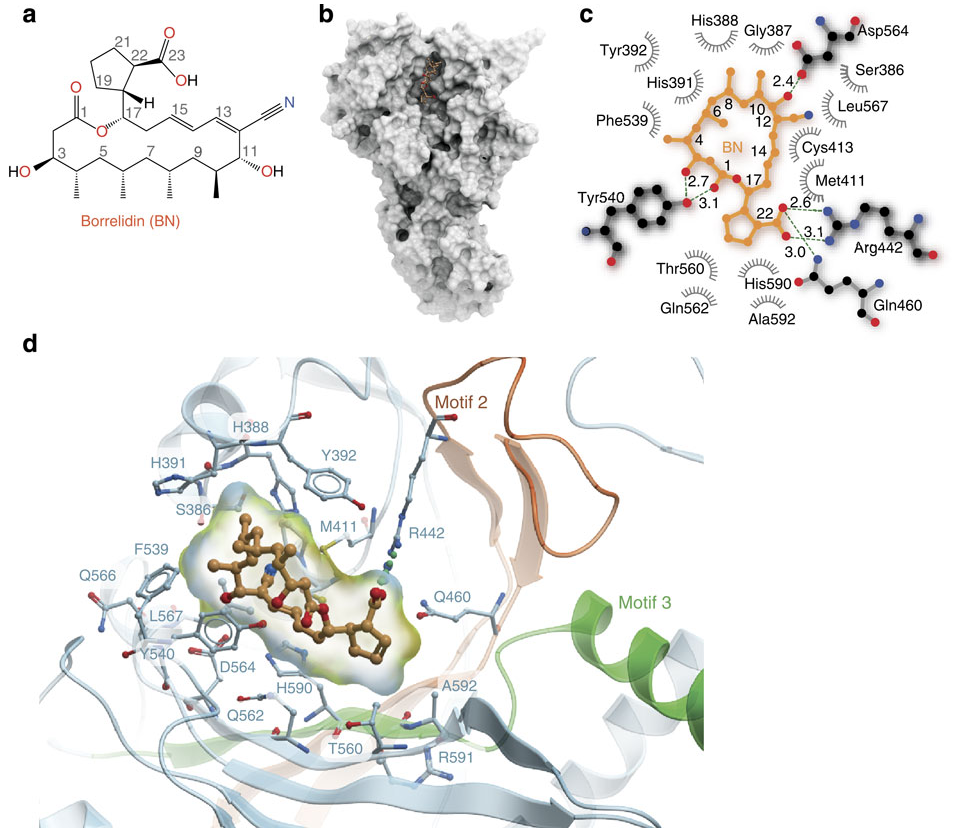 Figure from [Fang et al.](http://www.nature.com/ncomms/2015/150331/ncomms7402/full/ncomms7402.html)
Figure from [Fang et al.](http://www.nature.com/ncomms/2015/150331/ncomms7402/full/ncomms7402.html)
A professor in graduate school used to exclaim that “we all [cellular and molecular biologists] secretly want to be structural biologists”. At the time, as a structural biologist, I couldn’t help but agree. His exclamations were almost always prompted in journal clubs or lab meetings when an attempts to link a phenomenon of interest failed to make it all the way down the reductionist chain to the molecule of interest and the role a particular mutation may have played in a target protein’s behavior. He was implying that everybody yearned for the powerful finality that a picture of their molecule could yield.
I bring this up because I’ve just run across a beautiful paper - both scientifically and visually, the first figure of which is above. (paper here) This figure is a series of panels showing the natural product borrelidin and its interactions with it’s cellular target, the tRNA synthetase for threonine. And what does it show? It quite literally shows how these two molecules touch one another.
- a. The top left is a standard organic chemistry depiction of borrelidin.
- b. The middle shows borrelidin nestled into a groove formed by the surface of ThrRS
- c. Shows a schematic of the salient contacts between borrelidin and ThrRS and
- d. Is a zoomed in 3D depiction of the complex.
These pictures are nothing short of stunning. These are the sort of pictures that are indeed worth a thousand words; they capture and display some essential features of borrelidin: that it binds in a cleft in a particular way; that these bindings are mediated by physically matching and complementary aspects of the small molecule and the target protein. It is, indeed, no wonder that everyone wants to be a structural biologist - the pictures are just too good.
Although a,b,and d are relatively common in the field, I enjoyed seeing panel c which is a new visualization I haven’t previously spotted in the wild. It looks like like the latest from Open Eye which aims to boil down all of the essential information available in panel d. to a flat 2D image. This is the sort of information that most biologists and chemists looking at a protein-small molecule complex will be drawn to: which residues form the protein contacts with the small molecule? Which ones make hydrogen bonds? Which hydrophobic contacts? Where are the waters? For the purpose of understanding the essential contacts this is a slick tool. Although any structural biologist worth their salt will want to zoom around the structure in 3D, anyone who has spent time actually zooming around realizes how easy it is to get lost/disoriented. Flattening down the interactions to 2D and providing all of the essential information is fantastic - and I can image that med-chem guys doing lots of co-crystallizations will love this tool.
I should mention, raving as I am about the visual quality of this paper, that there is a pejorative view of structural biology as well: that these are just pretty pictures that add little-to-no value to the hard-earned insights of the molecular and cellular biologists whose work preceded them. And if these structures don’t result in hypothesis generation, then indeed they are just that. In this particular paper, the rest of the work is devoted to how borrelidin’s binding in this location results in the blocking ThrRS’s catalytic activity using structure guided mutagenesis and some straightforward enzymology.
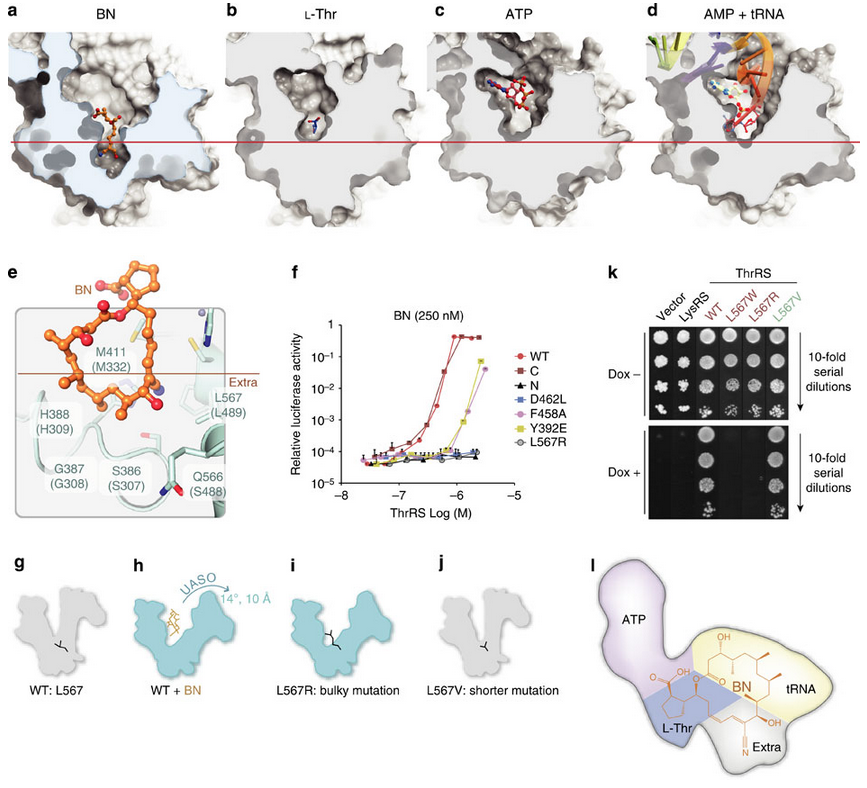
In the fourth figure (below), after demonstrating that borrelidin can compete for binding against the native substrates of ThrRS, the authors propose a mechanism for its action. Using co-crystals with borrelidin and the native structures they notice that borrelidin (a), and not the native substrates (b,c,d), goes deeper into the binding cavity and causes ThrRS to hinge-open. Is that how borrelidin works? Well they can recapitulate the hinge-effect of borrelidin through mutations in the hinge site (L567R) both enzymatically (panel f) and organismically (panel k), confirming that this is what borrelidin is doing. Not only is borrelidin directly occupying much of the active site; it is also changing the active site conformation to make it unfavorable for native substrates to bind.
As someone who have been involved in both structural biology and natural products I find this to be a delightful and fascinating approach to elucidating how these natural chemicals act so potently and exquisitely upon their targets. Kudos to the Guo Lab for their nice work and excellent visuals.