PLM-Local
Note: this is cross posted from ferritin
PLM-Local
This is a quick update post.
- Release of ESMC
- Began to port ESMC model in subcrate ferritin-esm
- Ported the model in this PR, and this PR
- However, when I went to implement the tokenizer, I ended up encountering some issues. Although ESMC is similar to ESM2 in that it is trained only on sequences, it shares a common codebase with ESM3 which uses structural information
- The major implication is that the tokenizer is not as simple and self-contained, and porting requires either:
- a full minimal reimplementation of ESM functionality or
- a simpler tokenization scheme that deviates from the current codebase
- While brainstorming on ESMC, I remembered that I hadn’t implemented METAL for AMPLIFY; so I did
- This makes AMPLIFY really pleasantly fast!
- I was therefore inspired to build and serve the binary via a new project, plm-local, and can build and deploy to a Conda channel
- h/t to luizirber for showing me this nice trick
pixi exec -c https://repo.prefix.dev/protein-language-local-public \
plm-local --protein-string MSVVGIDLGFQSCYVAVARAGGIETIANEYSDRCTPACISFGPKNR
# that's pretty awesome.
time pixi exec ... <as above> 0.53s user 0.65s system 42% cpu 2.800 total
time pixi exec ... <as above> 0.53s user 0.27s system 172% cpu 0.464 total
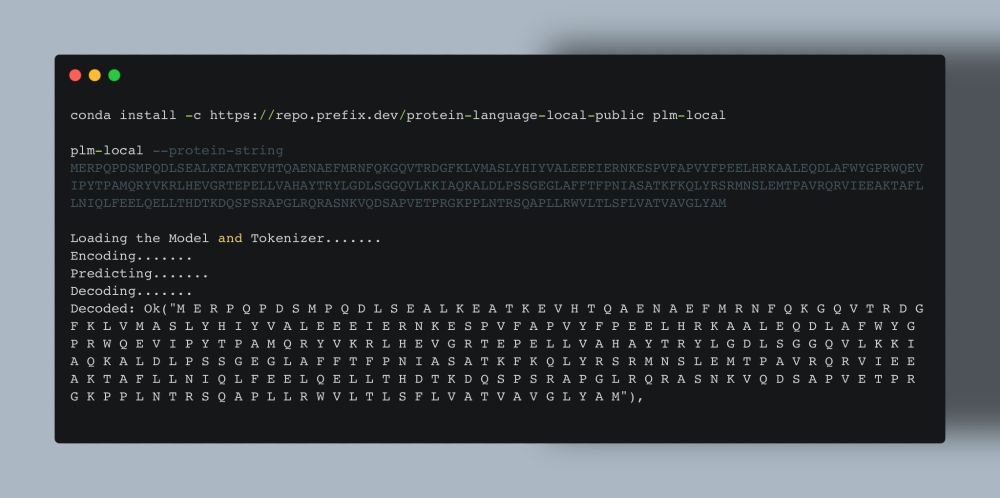
What it looks like at the moment.#
Current CLI simply encodes and decodes the protein. But it can do it in half a second with no compilation step or dependencies! Boom.