Ferritin Init
Note: this is crossposted to Github
Intro to Ferritin
This is an introduction to Ferritin, a set of Rust crates for working with proteins. At the moment ferritin is very much in alpha stage but I am pleased with the current progress and wanted to give a high level overview of the project as it currently stands.
At the moment I have in place what I think to be a reasonably well architected core data structure, an AtomCollection struct that is inspired by how pymol and biotite internally represent atomic information.
Most of the current and future features should be able to extend functionality via traits/methods while hopefully presenting s relatively small API that, because it is built on Rust, is nonetheless performant and portable. Additionally I have a few nascent crates that 1) handle inputs from pymol via it’s PSE binary file, 2) an initial implementation of a Rust version of molviewspec that can export json and/or self-contained HTML, and 3) an early set of visualizations built on top of Bevy.
In this post I will give a high level intro to the crates in this repo, a brief experience report of using Rust, and glimpse of the next steps.
// current layout of Atom Collection.
pub struct AtomCollection {
size: usize,
coords: Vec<[f32; 3]>,
res_ids: Vec<i32>,
res_names: Vec<String>,
is_hetero: Vec<bool>,
elements: Vec<Element>,
atom_names: Vec<String>,
chain_ids: Vec<String>,
bonds: Option<Vec<Bond>>,
}
Molecular Visualization#
In the Rust world, Bevy seems to be ascendant with a lot of activity on it’s Discord channel.
I stubbed out an initial vizualizer using pdbtbx
and that worked okay but I realized that I would want to have a set of ergonomic functions
for working with stucture files that would allow me to generate new subsets of atoms and new
visuals. I also ran into the issue that pdbtbx doesn’t parse/create bonds. However, with AtomCollection
I was able to copy biotite’s bond creation algorithm (using lookups from CCD) and now was can
relatively easily create bonds as well. So once AtomCollection was implemented I ported that code here. At the moment theres
over and implemented a ball-and-stick viz.
Viz is clearly a growht areas especially since I think I should be able to mimic the hierarchical nature of selction/visualizations that Molstar has done such a great job on. And Bevy will allow you to extend/animate etc.
// simple examples
cd ferritin-bevy
cargo run --example basic_spheres
cargo run --example basic_ballandstick
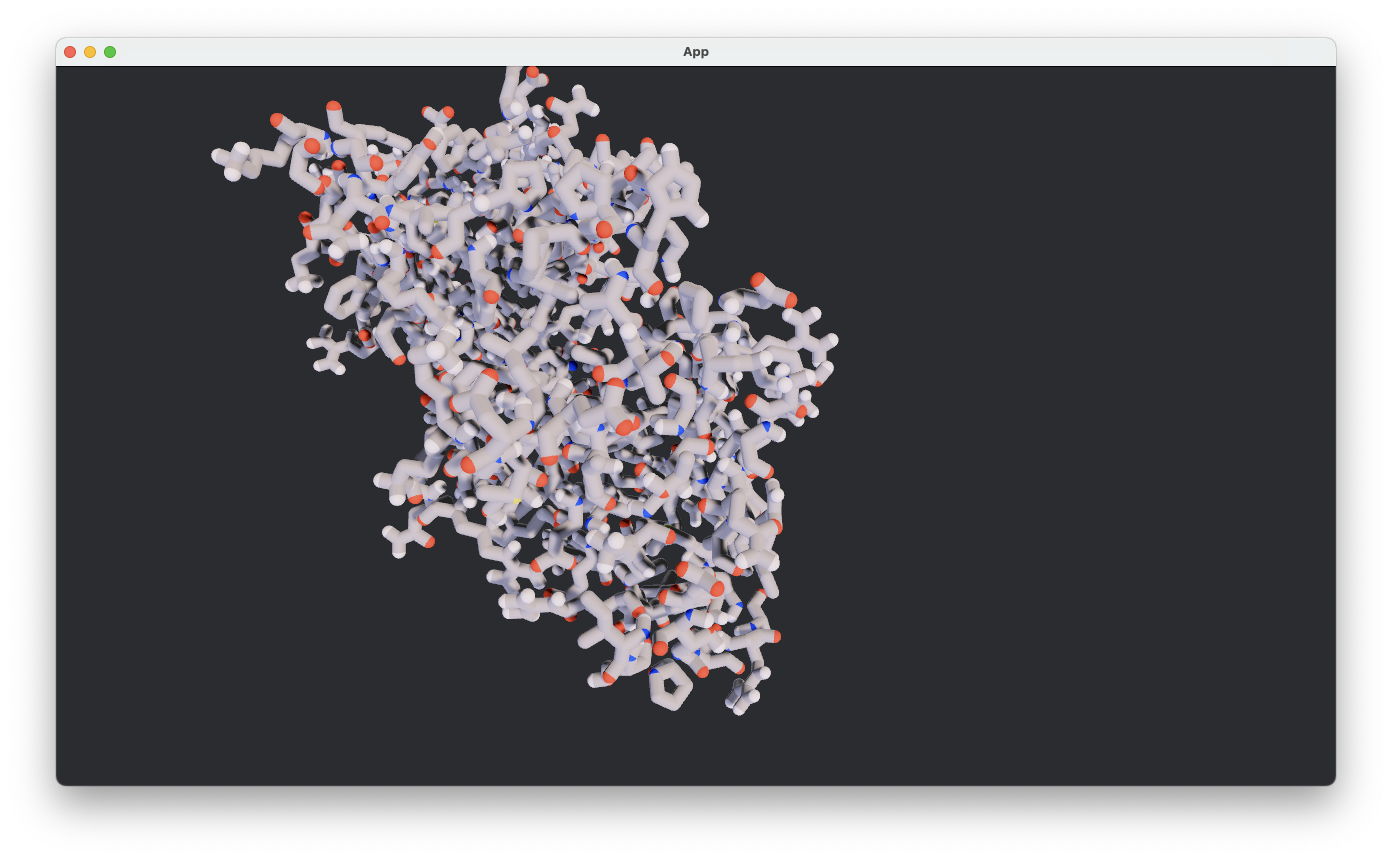
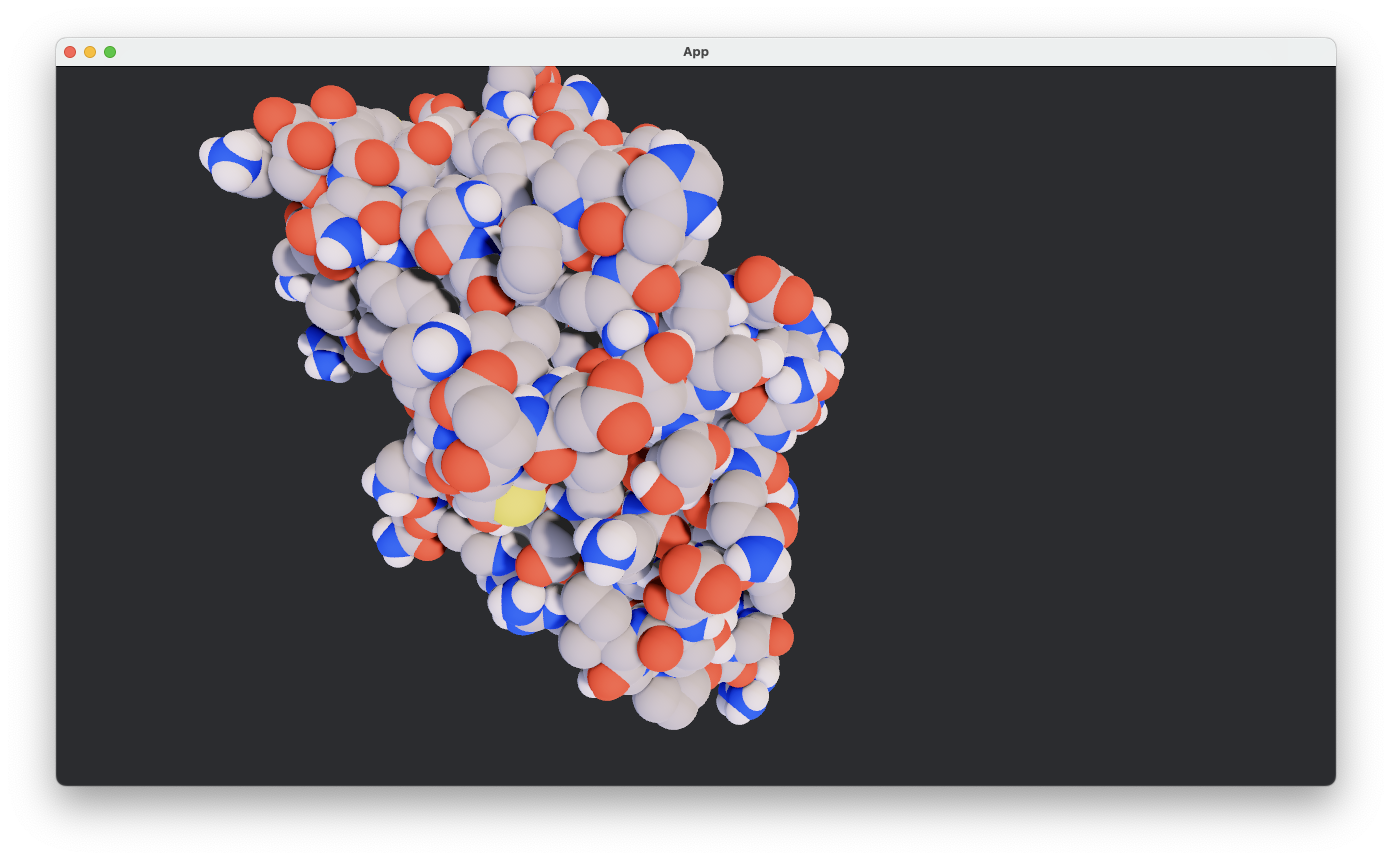
Pymol PSE Loading#
Pymol is the lingua franca of protein structure analysis. It is open source, full of utilities and what many structure biologists are raised on. My initial interest in the pymol PSE format was the idea/hope that I would be able to create a workflow where I could use pymols extensive utilites but then serialize to the web using molviewspec. Somethigng like this:
# inital impetus for Rust serialization library
analysis in pymol -> save to pse -> serialize to molview spec
During the creation of my initial attempt, pseutils, I
ended up learning a few things and abandoning the approach. The first thing I learned was the power
of serde_pickle, a library that serializes binary pickle
data. Using this and a bit of pymol code spelunking I was able to mostly recreate in Rust structs the pymol
PSE data. Form this I learned quite a bit about how the high level python API and the low-level C++ implementation
have diverged over the years - there are lots of steps of indirection between the pymol API and the low
level data format (or at least it seemed so to me). One of the lessons I learned was that we should be able to
use a much more direct relationship between low level represetnation and high level API (more on that below). The
second thing I realized is that I would need to spend a lot of time recreating the high level intent of a pymol
command from its low level bits - e.g. “color by helix with rainbow” would need to be recreated from looking
at each atom. I very much preferred the molviewspec style which is much more like React: data->function->viz.
// the PSEData struct is the entrypoint to Pymol's binary `pse` format
use ferritin_pymol::PSEData;
// all pymol pse data available herein
let psedata: PSEData = PSEData::load("tests/data/example.pse").unwrap();
Serialize to MolviewSpec#
I’ve recreated the MolViewSpec hierarchy in rust. We should then be able to translate our structure, selection/component, and representation data from any source and export it as molviewspec-json (MVSJ). As I was initially interested in creating a pymol converter I did make a utility in that would generate a self-contained HTML page with accompanying MVSJ json file.
// cli to convert pse.
ferritin-pymol
--psefile docs/examples/example.pse \
--outputdir docs/examples/example
// Example builder code to generate molviewspec json
// Components are selections...
let component = ComponentSelector::default();
// Representations need a type. I implemented only one or two
let cartoon_type = RepresentationTypeT::Cartoon;
let mut state = State::new();
state
.download("https://files.wwpdb.org/download/1cbs.cif")
.expect("Create a Download node with a URL")
.parse(structfile)
.expect("Parseable option")
.assembly_structure(structparams)
.expect("a set of Structure options")
.component(component)
.expect("defined a valid component")
.representation(cartoon_type);
Ferritin-Core#
As mentioned, the three crates above were originally two standalone projects: pseutils, and protein-render; these are now archived. I had also been playing around with visualization in Blender. All together I realized I was fighting or working around different APIs to do what I needed. And so I decided - why not try writing the API that I wanted. Okay so what do I want?
Simple data structure#
I wanted a simple data structure that is maximally flexible and that I can work off of. One interesting thing poking around the pymol internals
is that the underlying binary representation of things is computer optimized using a Struct-of-Array
style that keeps the coordinates tightly packed in memory. Below is some code from the ferritin-core crate that points this out.
// the binary layout of pymol's PSE files
// uses a Stuct-of-Arrays approach to memory layout
pub struct PyObjectMolecule {
pub object: PyObject,
pub n_cset: i32,
pub n_bond: i32,
pub n_atom: i32,
pub coord_set: Vec<CoordSet>, <--- Coordinates
pub bond: Vec<Bond>,
pub atom: Vec<AtomInfo>, <--- AtomInfo
....
}
// note heterogenous data especially Strings
pub struct AtomInfo {
pub resv: i32,
pub chain: String,
pub alt: String,
pub resi: String,
pub segi: String,
...
}
// all numeric. some indexing and then the entire coordinate set
pub struct CoordSet {
pub n_index: i32, // 1519
n_at_index: i32, // 1519
pub coord: Vec<f32>, // 4556 ( 1519*3 ) <---- [x,y,z,x,y,z....]
pub idx_to_atm: Vec<i32>, // 1 - 1518
}
It was interesting to me that a recent and growing python package, biotite, has a similar organization style in its use of AtomArrays
to store atom info in Numpy Arrays. I’d also watched Andrew Kelly’s talk on data oriented design which is a strong pitch for
making software fast by paying attention to it’s memory layout. Sounds good. After googling around a bit I saw a few Struct-of-Array libraries but it seems
that most people just kind of roll their own - e.g. use vectors of homogenous data in their struct types and handle the index themselves. Thats what
AtomCollection ended up being.
Thats Fast … and also ergonomic.#
I think this library should be relatively quick if I end up using packed data, use iteration, and avoid copying. At least thats what the Rust world seems to indicate. We’ll see. One other thing I wanted is something simple and ergonomic. If you read the code in biotite one can’t help but be impressed at how well organized, cohesive and simple the code is. I contrast this with the pymol code which has acquired multiple decades of indirections and lots and lots of indirections. So the plan is to try to keep the high level API smooth and nice and hide away any complexity while keeping the speed up. I think its possible and if so, it might turn out be be widely useful.
At the moment I have basic IO via the excellent pdbtbx crate; selections and filtering of residues; iteration through residues
using an indexing mechanism based on iterators; and a set of tests indicating initial good behavior. Theres lots of work to do but I think
the core AtomCollection can now be reasonably extended via new traits/methods fairly easily.
// the one ring
use ferritin-core::AtomCollection;
// utility to load a stet file
let ac: AtomCollection = get_atom_container();
// iter_residues_aminoacid over residues using indexing/slicing
// `collect` invokes a copy
//
// this look very pythonic to me but should run at C speeds
let amino_acids = ac.iter_residues_aminoacid().collect();
Rust Experience Report
Learning Rust#
Its been a bit of learning curve but I’m glad to be working in Rust. Coming from a dynamic background (R/Python mostly) and static language can be tricky and Rust especially so. But with help from Claude+ZED, I was able to get over the learning curve and now feel like I’ve got some momentum.
Some tough bits:
- Lifetimes are still far from intuitive but I didn’t come across them for awhile. And when I did hit them my LLM friends were there to help.
- Code organization. All languages have their quirks about the best way to organize your files/functions/structs. Rust is no different. I did download RustRover at some point when I wanted to move some files around and make sure all the references are preserves. Would love to see that end up in Zed at some point.
- Occasional rust language server insane memory/cpu use. Not sure how to avoid those but they require foce-quit and restart
Some Wonderful Bits:
- Iterators and clojures are really great and I feel comforatable there.
- When I am working within methods using iterators I feel really good and I feel that the language is working for me. (they are handling ownership, etc)
- Structs/Methods. As in the code organization, knowing when/how to give something its own identity didn’t come natural. I started programming in functions but ended up really liking associating methods with fewer structs as a way to minimize overhead.
- Traits. I found these a bit awkward to think about/use. When do I write them? when do I need them? I came to appreciate them at two occasions. At one point I needed to refactor some IO code and the trait boundary was the perfect way to define the bit of code to move. Similarly as my core Stuct accumulates methods, I will want to think about grouping these functions into traits and using these traits to define new extension behavior.
- Private by default. It can be frustrating but… it pays off. I had just been teaching myself a python library where there is a large data structure which gets heavily mutated and its hard to keep track o whats happening. If your fields are private by default it limits how the mutations happen but this mutability gives you a lot more confidence when looking at a small piece of the code that you are understanding all of the relevant bits. So not ergonomic but big-picture healthy.
- Autoformat. Love it. I don’t think about formatting at all except when I am tying to stash data in a struct. then i can flag the region with
#[rustfmt::skip]
Claude & Zed#
Both amazing and helpful tools. Zed is fast and I’ve been using it since it was released. The incorporation of LLM APIs has been superb. I use the chat window for exploring approaches or solving problems and I use the inline box for small fixes, alphabetizing methods, inline tweaks, and other small, local tasks. Combined they definitely keep you in flow.
Next Steps
Lots to do. But what I would like is:
- a decent interactive viewer in Bevy with common visualization (ribbon, cartoon etc).
- a wasm build of the above.
- some common protein utilities (e.g. rmsd)
- note: this is the best/easiest entrypoint for new potential contributors
- ML tokenizer crate. E.g. for use in DL models via candle
If any of this interests you - reach out!