Return to LigandMPNN
Note: this is cross posted from ferritin
Returning to LigandMPNN
The original motivating goal of this project was a pure-rust-WASM ProteinMPNN/LigandMPNN implementation for fast local design. In previous posts I discussed:
- Core Protein Data Structures for efficient protein representation using the Struct-of-Arrays style. link
- The LigandMPNN Trait to define functions for extracting features from a protein representation. link
- The Candle implementation of the Amplify model (parts 1, 2, and 3)
Now that I was able to get my feet wet on a protein language model implementation, I am ready to return to the more architecturally challenging problem of {Protein/Ligand}-MPNN. This post will describe a few of the challenges faced in porting that library over and the current state of the model.
Issues to Solve.
Model Complexity#
There are a few differences between Candle and PyTorch in terms of how:
- they handle dimensions selection (PyTorch: NUMPY-like; Candle: methods like
i,narrow,squeezeandunsqueeze) - whether the Tensors can be mutated in place (PyTorch: yes; Candle: no)
- specifying matrix contiguity. (PyTorch: ?; Candle:
.contiguous())
As a ballpark approximation we can take a look at a few of the function call types that handle those differences. I am
calculating the occurrences of function calls in the code bases here and here and showing
the aggregated results in the table below. The LigandMPNN data is inflated because these calls also include the sc.py file which allows for side-chain packing. Nonetheless,
from the perspective of implementation, it should be clear that the potentially tricky implementation bits in LigandMPNN are far greater than in Amplify.
# ligandMPNN or huggingface AMPLIFY_120M dirs
rg -c '\[' *py
rg -c 'gather' *py
rg -c 'scatter' *py
rg -c 'mul' *py
| Term | LigandMPNN | AMPLIFY_120M |
|---|---|---|
[ |
910 | 12 |
gather |
55 | 0 |
scatter |
5 | 0 |
mul |
52 | 3 |
Model Loading#
In working with AMPLIFY, one of the key successes was being able to load Amplify’s model into a VarBuilder using the
from_mmaped_safetensors. You can then build your model by accessing the Tensors by name. It allows you to match the PyTorch model using layers that make sense by name. During this process,
I noticed that there is a similar function for PyTorch files - from_pth. Excellent! This gives me a new tool I lacked - the ability to load the model with the exact same
names as the PyTorch model. I should be able to load this file and have all the Tensors match and use/account for all layers. It turned out there was a hiccup in that the PTH
and safetensor formats differ a bit and you need to be able to access the PyTorch statemap. I submitted a fix here and began
assembling the model into my pre-existing code. This is where I began to run into a number of issues related to Tensor dimension.
Dimension Matching#
As I began to load the Tensors in from the PyTorch file, I began to hit errors introduced by the incompatible syntax mentioned above. In this case I would need to compare the PyTorch code with my Rust code and 1) introduce the Candle syntax while 2) maintaining the flow/intention of the model. For this I leaned quite heavily on Claude/Sonnet3.5 via the Zed editor. This was an invaluable experience and further impressed me as to LLM capability. Here is a taste of Claude’s explanatory power; full gist here

Speed#
After a bit of work I was able to load the model and run it where run means execute the model with an input and get an output without failing. My strategy had always been to get it
running then get it to pass tests so I was pretty pleased. However, the model took minutes to run! Not what I was looking for. So after pinging on the Candle Discord, I realized
that I was on MacOS but that I had been using Device::CPU. What if we switch to Device::Metal? I had to rework the code a bit to get the Device passed in but then I hit a bunch of errors like:
// Metal doesn't support 64 bits!
Err(WithBacktrace { inner: Msg("Metal strided to_dtype F64 F32 not implemented")
// No gather calls on integers!
Err(WithBacktrace { inner: Msg("Metal gather U32 I64 not implemented"),
// No scatter-add on integers!
Err(Metal(UnexpectedDType { msg: "scatter-add ids should be u8/u32/i64", expected: U32, got: U32 }))
The first fix is to convert a number of F64/I64s to F32/U32. I then needed to track down and implement a few 2-line additions to Candle’s Metal kernels that would allow the kernels to work. These were the PRs for Gather and Scatter Add. The result was impressive. My initial model ran in 3 minutes; the new model in 8 seconds! Okay, we can work with that.
cargo instruments -t time \
--bin ferritin-featurizers \
-- run --seed 111 \
--pdb-path ferritin-test-data/data/structures/1bc8.cif \
--model-type protein_mpnn --out-folder testout
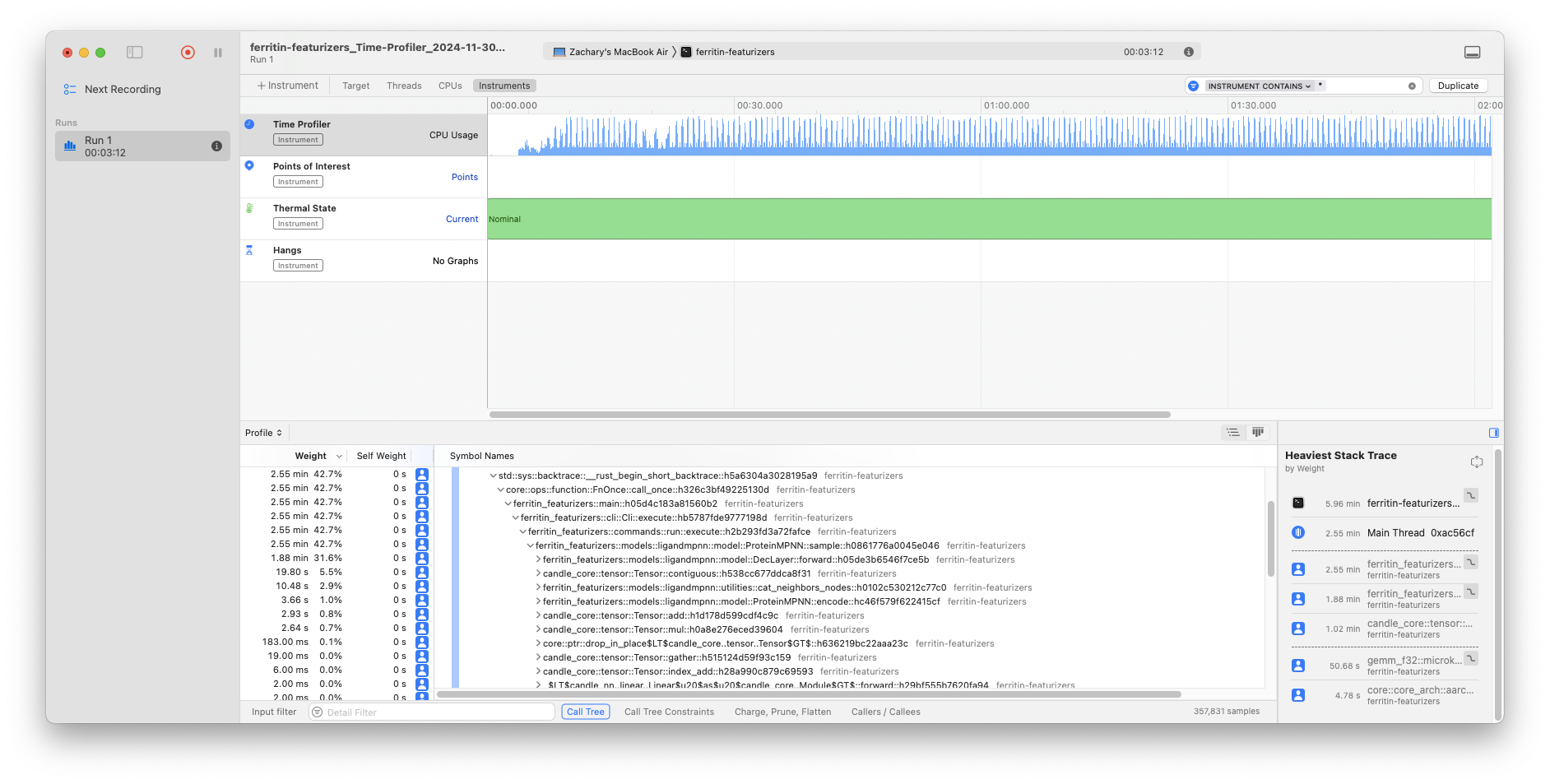

Testing Suite.#
I’ve started a test suite to match LigandMPNNs and have begun implementing the CLI code for it. As of Today, December 2 there is not much to show. But I am satisfied with where the project has gotten and am impressed by Justas Dauparas and his collaborators on this implementation. There are still some hard bits ahead.