Foray into Molecular Nodes
TL;DR: Wobble is a Python cli Library that can currently be used to generate an animation where residues wiggle based on a PDB’s B-factor. It is a proof of principle library demonstrating one way in which we might use Blender and a rendering engine for fun/interesting protein visualizations.
Background#
Protein animations are awesome and I’ve been interested in them for a long time (see PCR Rap below). And in the last month or so a few things rekindled the interest: I saw the absolutely beautiful Evolutionary Scale website and I attended a local meetup at VantAI where Brady Johnson gave a walkthrough of Molecular Nodes. It was pretty clear that a skilled animator could replicate each of the EvoScale animations using Molecular Nodes and watching how Brady operated inspired me to take a crack at a few visualizations that I had wanted to make in the past but where I thought a dynamic environment like Blender would be really beneficial:
- Mapping sequence conservation onto a representative structures and having it animate. (some progress here)
- Creating a standard tool to generate protein-ligand interactions where the ligand is emitting light. ( Brady showed how to do this but I didn’t implement this)
- Create a set of vizualizations that represent residues or inter-actomic interactions - e.g. for things like NOESY spectra, cross-correlation matrices, or other correlation matrices.
Also - even though I am interested in making images, I am particularly interested in making reproducible images that I can apply to future projects.
Blender & Molecular Nodes#
Interactive Dev#
When I began this project I was hoping that I would be able to use some sort of REPL-style development where I could make changes to code in my editor and execute them to see what happens in Blender. I thought that I would be able to build up my networks programmatically or maybe even declaratively as a nested JSOn tree or something. But I ran into some issues.
- Blender ships its own python version and its internal repl is not available to you.
- There are tools to open blender’s python to your Editor. I tried a few routes and found them brittle.
- I discovered basilisp-blender which is aweosme and works great. I even generated a number of functions in basilisp but ultimately decided against it because even though the REPL experience is really nice/good - I ultimateyl want to be able to distribute reusable code to python users and this adds an extra toolchain.
- I discovered that you can delete Blender’s python and that it will then look for a new python env.
# in my case deleting the Blender python leads to using my env
# see https://github.com/zachcp/molnodes-utils/blob/main/pixi.toml for my dev setup
rm -rf /Applications/Blender.app/Contents/Resources/python
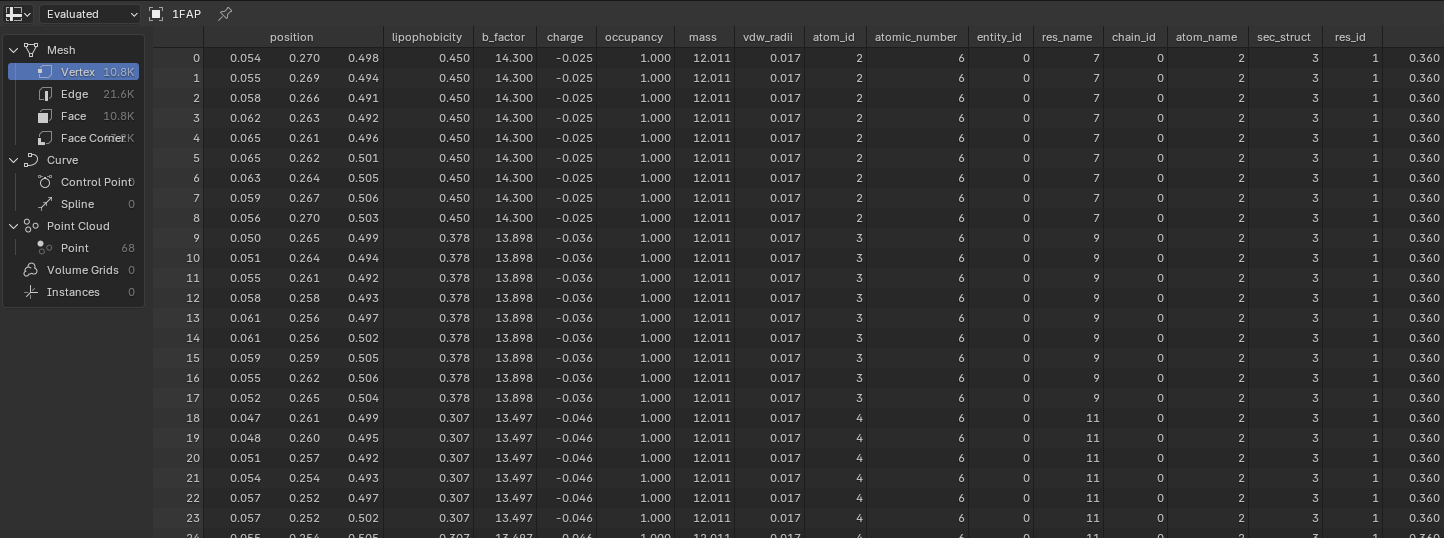
Saving and Accessing Data#
MolecularNodes has a few entry points for creating molecules. Ultimately, however, they become a Molecule which instantiates a Blender object with the
create_object method. This will register the data
in bpy.data.objects. That data can be accessed via the api: bpy.data.objects.get('1FAP') or viewed in the Spreadsheet view.
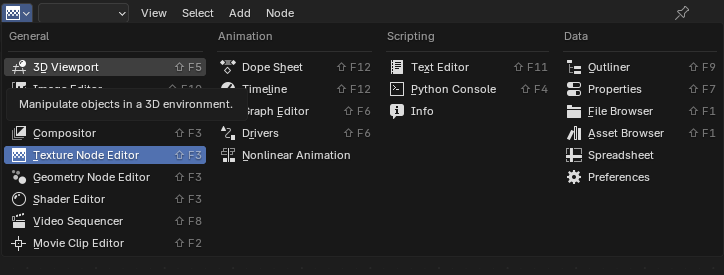
You can access the spreadsheet view, along with all the other view types using the drop down in every window:
Blender Ops#
When MolecularNodes loads up, it also attaches a branded set of Nodes to your object. These are GeometryNodes
that Brady has put together. In the GeometryNodeEditor you can click on a node and if you hit Tab you will “step-into” the node group ( if it is a group of nodes). If you do this you can
quickly begin to see that MolecularNodes is turtles nodes all the way down. I show a few of these below.
Its Nodes All the Way Down#
Default MolecularNodes
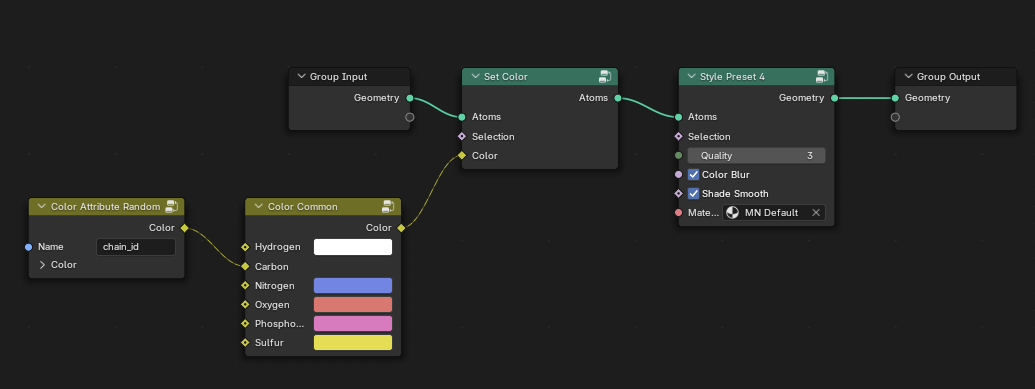
Is Peptide?
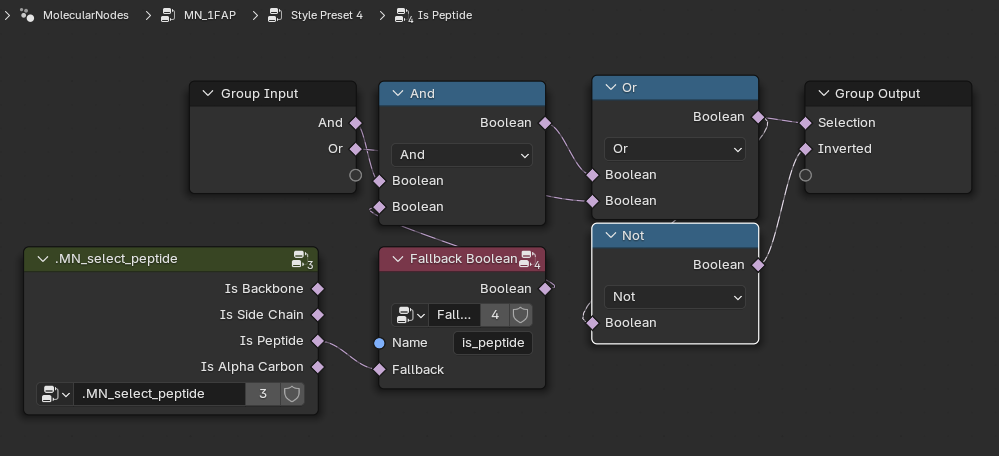
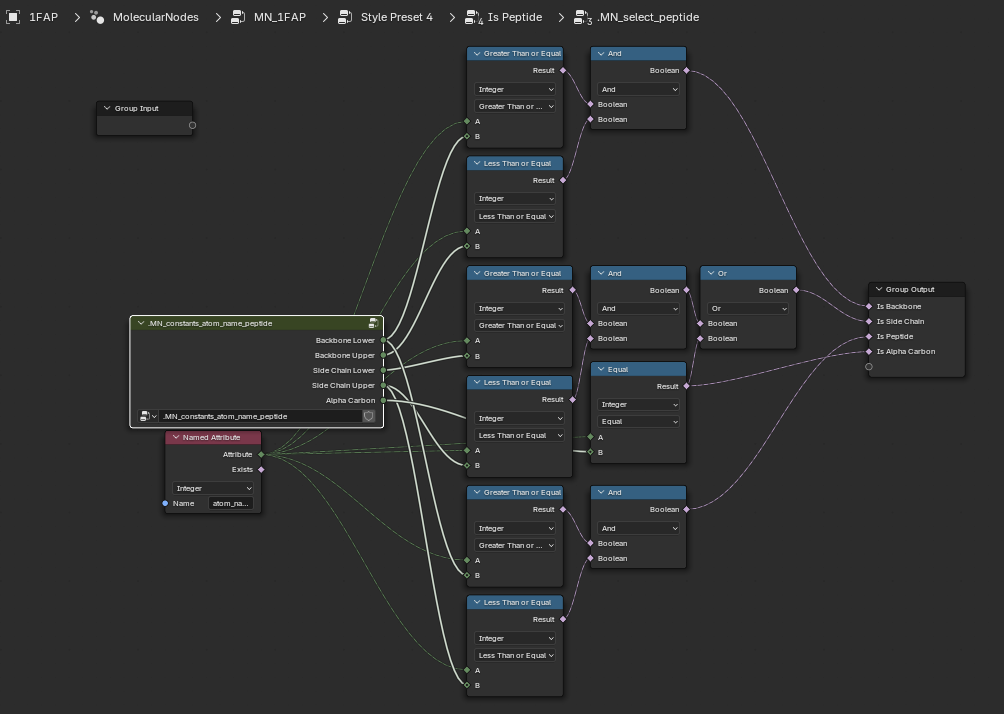
Select Peptide?
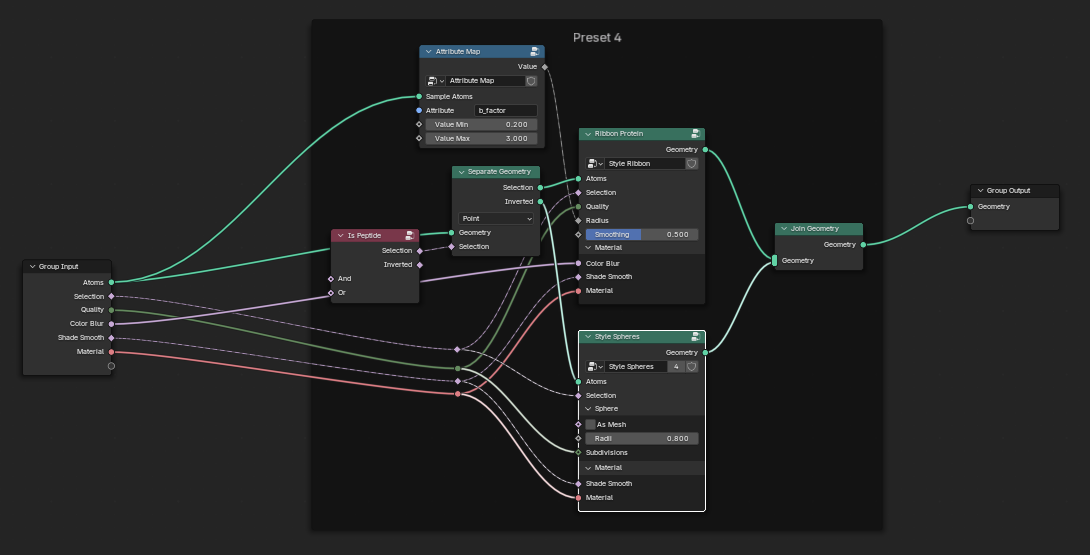
Preset 4
Convert ALL-THE-NODES to Python?#
So the tangle of nodes we see above makes the tractability of my original approach - e.g. building up a declarative tree, a la molviewspec, a bit less attractive - it will be a lot of code and bookkeeping. But what if we just autoconvert these to python and see what happens…
I tried it but ultimately was unsatisfied. If you are interested here is the conversion script that will iterate through Brady’s nodes and use Node2Python code to generate python classes for each of the Molecular Nodes. However, this results in many, large files. This is not obviously better…..
My Strategy for Wobble#
Lets take the simplest path that gets me where I want to go without reinventing the wheel from whole cloth. That seemed to me to be the following:
1. Open Blender and Load up the Network Editor
2. Play with Network IN THE BLENDER GUI until things look ok.
3. Save the NodeGroup as a python class using the `NodetoPython` plugin.
4. Move the output class to a new package, `wobble`.
5. Create helper functions to load the PDB and swap the NodeTree
6. Write a script to render the images.
Pros#
- it works. I can distribute it as code and program against it.
- I can do the same again and again and register each class as a file that applies styles to a protein and allow that set of objects to be treated like any Blender mesh.
- allows you to use many tools like Blendersynth to create animations
Cons#
- the
wiggleclass is 6900 lines of code. Its the entire node tree….. - thats kind of crazy.
Future Directions#
My Original Goals#
I didn’t get any of the 3 original goals to completion but I am in a much better place to do. A brief comment on what would be needed:
Mapping sequence conservation onto a representative structures and having it animate.
Right now I am using the PDB loader. I would need to find/implement a PDB->Object function and do the conservation-to-B-factor-mapping upstream
Creating a standard tool to generate protein-ligand interactions where the ligand is emitting light.
Doable. I like this one because its beautiful and simple and hard to do with other programs. Would require setting up a reasonable node tree to separate out proteins form ligand and add light emitting properties to the ligands.
Create a set of visualizations that represent residues or inter-atomic interactions.
This is a bigger lift as it would require a separate data structure and a set of nodes to map them together. Not hard to do in a programming language but would need some research for how to proceed within blender.
General Moleculear Nodes thoughts#
After playing around with Blender/MolecularNodes for a bit I think there are a few areas where it could fit/excel in the ecosystem.
- The advantages of Blender are clear: animation, textures, lighting etc.
- Blender distributes
bpywheels so you can EASILY use Blender via the CLI. bpy - You can basically do anything with the API.
- But…. theres a lot of messy state to handle.
- People love using Brady’s tools and building on top.
- The main mode of development SEEMS to be:
GUI-->blendfile - Blendfiles are not bad but not great for programmatic access.
- I show one route to generating tools programmatically via
GUI-->python
In general, I don’t think you will ever fully get away from the GUI because thats where you will need to live
if you want to make complicated, bespoke graphics. However, I think there is a lot of territory for potential improvement
- ideally there would be something like molviewspec that lets you programmatically build up a scene and then emit to a blender environment. This would be more flexible than what I have above (e.g. totally pre-canned) but would let you have a lot more programmatic flexibility in treating Blender as a rendering tool and not an ecosystem. But… it would leave open the possibility of entering the Blender ecosystem for all of its goodies.
I think it would take some brainstorming and some trial-and-error to find a pattern that works - maybe it would be good to chat with the molstar/molviewspec team to see how they have approached the backend architectures they have worked on.
Bonus: PCR Rap.#
This was one of the first animations I did. Lots of printing pml commands…
Original Animation#
- Code here
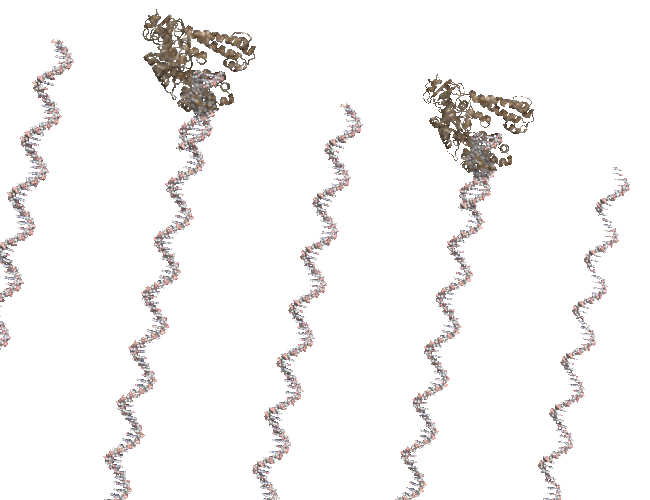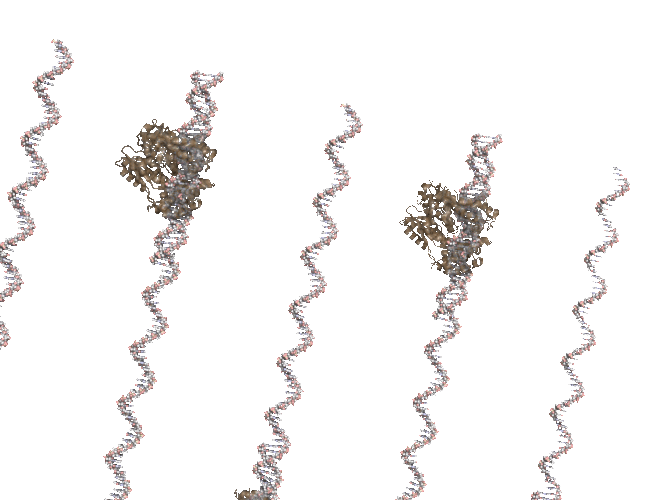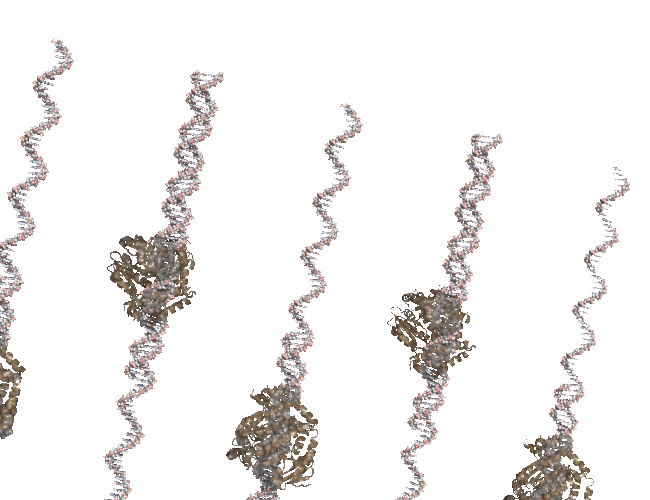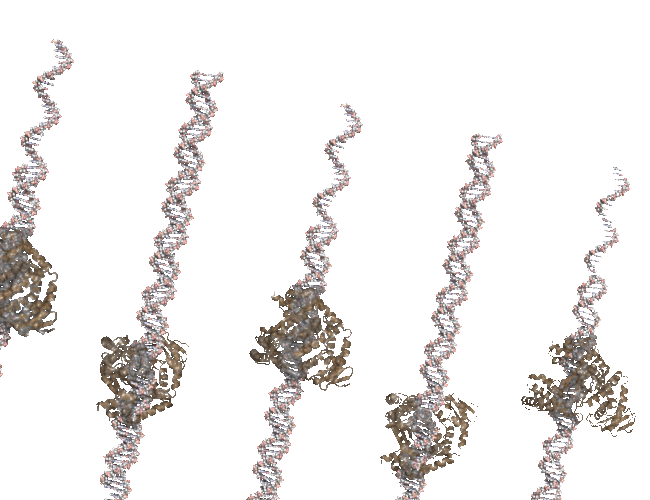
You Tube Video#
Links#
- wobble package under discussion
- node2python tool to convert GeometryNodes to python code.
- pixi fantastic tool
- MolecularNodes fantastic tool
- basilisp clojure+python
- basilisp-blender creates a REPL in Blender you can connect to
- blender fantastic tool
- blendersynth simplifies generating images.
- molviewspec declarative, nested specificaiton for generating images
- pseutils recent project to turn pymol into molviewspec
- pcr_rap classic science rap
- pcr_code code to generate the polymerases
- evoscale
- vantai
- noesy