PCR Animation with Pymol
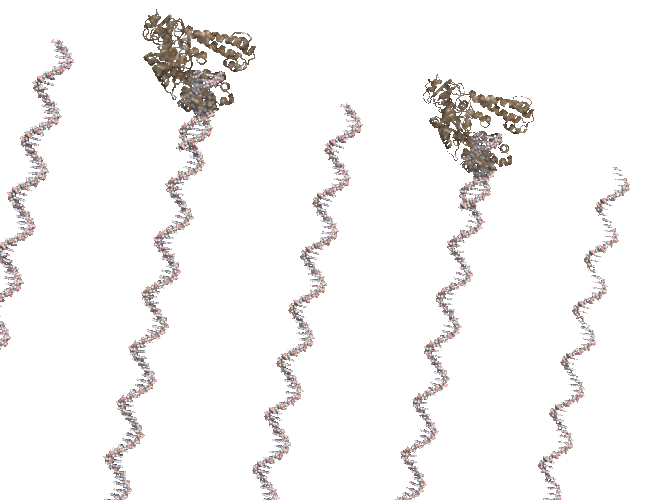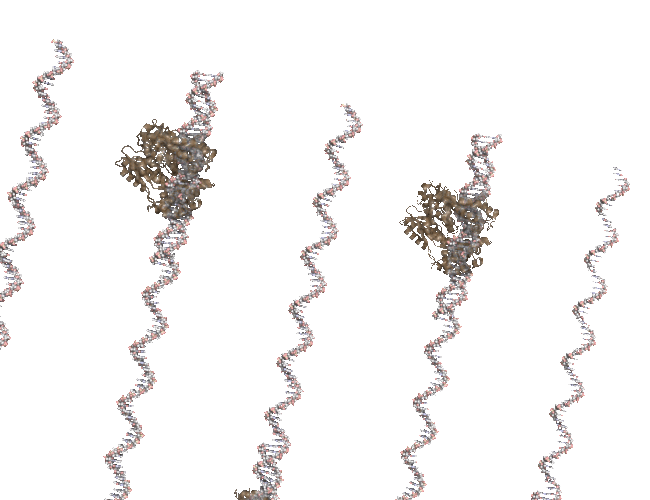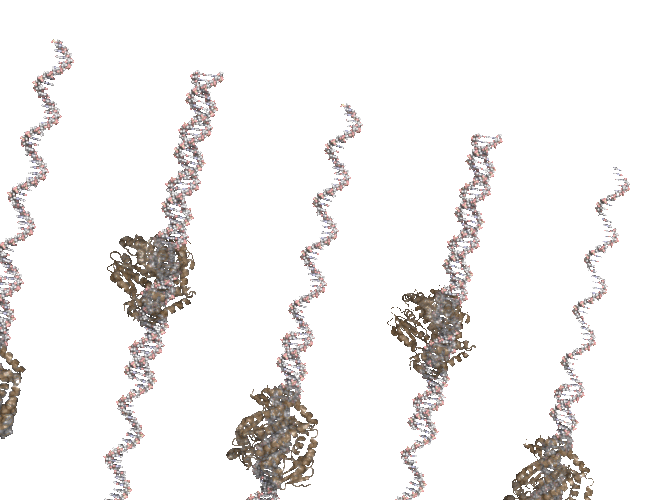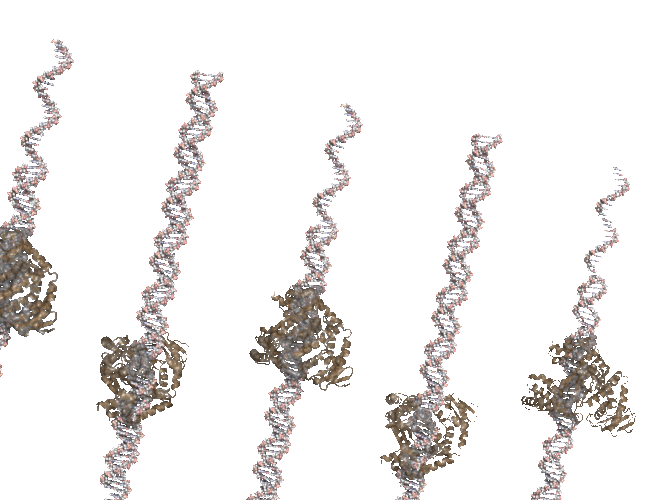
I was cleaning up some old files and uncovered the code I used to create a PCR-like animation for the PCR Rap and I’ve posted it to github.
The animation (seen above) is made in Pymol and uses a series of rotations and translations of a polymerase around a double stranded DNA. At the time I remember struggling with setting the view, coordinating the state of each polymerase and DNA strand and so instead of banging my head against PyMol I simple wrote a script that would write out the commands that I wold normally use in pymol and then run that script in pymol. it worked fine in the end but something like Quil/Processing is how I would do it if i was writing again. Quill interface to JMOL, anyone?
#a portion of the python file to calculate the rotations/translations
print "#molecule %s" % (y+1)
print "hide sticks,dna%sa" % (y+1)
print "show sticks, dna%sa and resi 1-%s" % (y+1, 15+x/2)
print "orient dna%sa" % (y+1)
print "translate [3.14,0,0], dna%sc; rotate x, 18, dna%sc" % (y+1,y+1)
...
print "png frame_matrix{:03d}.png, width={}, height={}".format( x+1, width, height)
# a portion of the pml file that was generated and was used to
# set the state each frame for each mol
# rotations were 18degres on around the x axis defined by orenting on
# the target mol
# translations were 3.14, the basepair distance
#molecule 1
hide sticks,dna1a
show sticks, dna1a and resi 1-40
orient dna1a
translate [3.14,0,0], dna1c; rotate x, 18, dna1c
#molecule 2
hide sticks,dna2a
show sticks, dna2a and resi 1-40
orient dna2a
translate [-3.14,0,0], dna2c; rotate x, -18, dna2c
set_view ...
png frame_matrix051.png, width=1200, height=800
My translation of 3.14 angstroms is about right but my startng position was off. You can see the DNA strands “running ahead” of the polymerase. The code is available in case anyone wants to tinker.