Chemdoodle Sketcher Gadget
This week I put together an interactive Shiny Gadget that wraps the Chemdoodle Sketcher JS. The basic operation is as follows.
# install the library.
# needs rJava with Java 7 or greater. see below if difficult to install
devtools::install_github("zachcp/chemdoodle")
# launches the chemdoodle sketcher and stores the resulting JSON
moljson <- drawMolecule()
# processes JSON to a CDK AtomContainer
mol <- processChemDoodleJson(moljson)
# convert to SMILES
smi <- toSmiles(mol)
The sketcher itself looks like this:
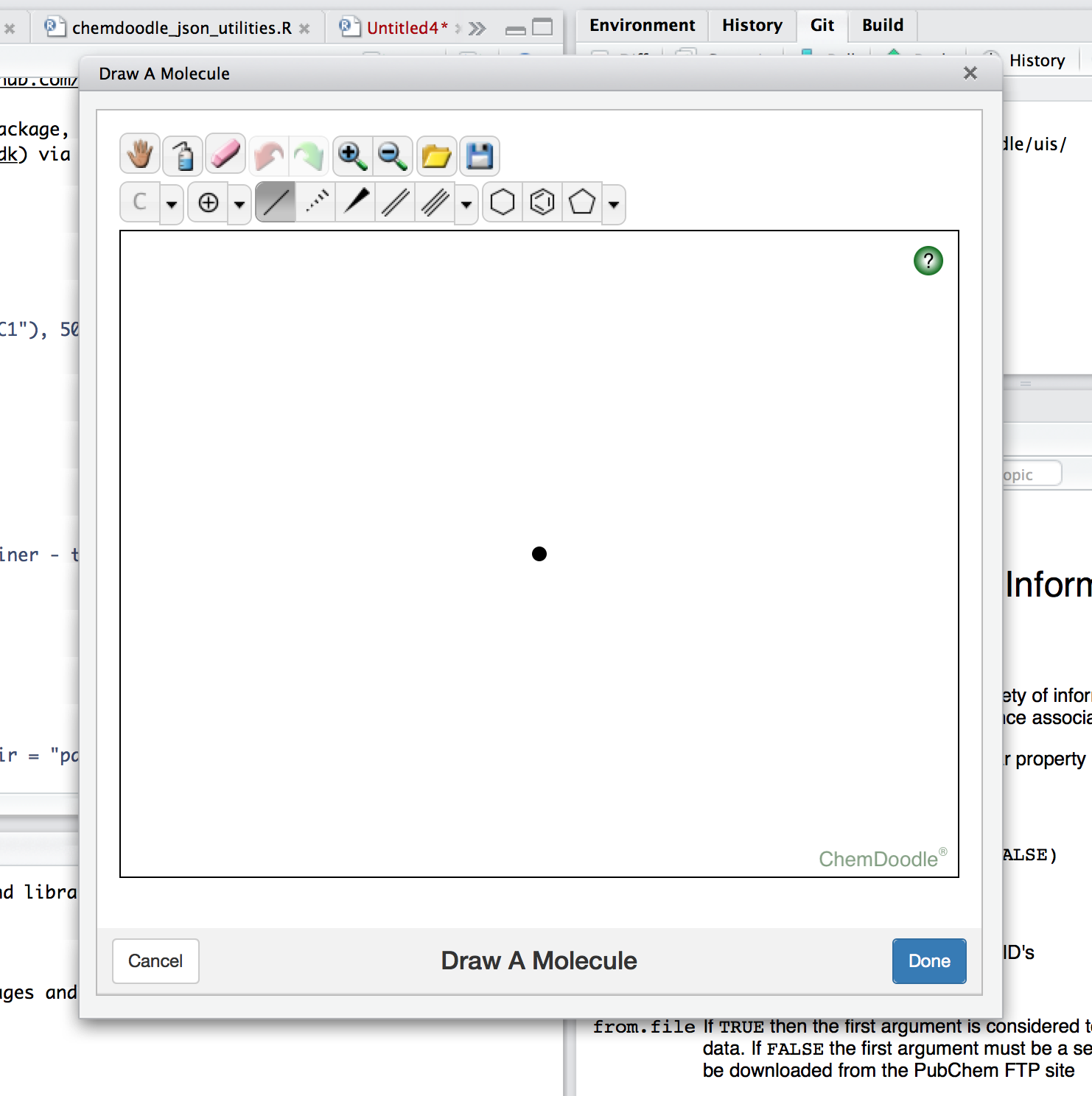
Heres a few technical thoughts on getting the plumbing to work.
The HTMLWidget#
The core tech is using the htmlwidgets package/pattern to instantiate a Chemdoodle WebComponent. This worked similarly to previous components with the exception that the Sketcher utility uses javascript to create the toolbar and does it in a way that destroys the div information needed by HTMLWidgets. To get around this one needs to supply a custom HTML function to the widget (h/t @timelyportfolio) that will render the widget and then assign it an id we can use.
#' @import htmltools
chemdoodle_sketcher_html <- function(id, style, class, ...){
tagList(
tags$div(
id = id,
class = class,
style = style,
tags$script(sprintf(
"
var sketcher = new ChemDoodle.SketcherCanvas(
'sketcher',
570, 440,
{useServices:false, oneMolecule:true}
);
//assign the sketcher using expando
// so that we can access it in the htmlwidget render
document.getElementById('%s').sketcher = sketcher;
"
,id))
)
)
}
The Shiny Gadget#
Once the htmlwidget is up and running you want to be able to pull values back from it into R. Fortunately the devs at RStudio
have been hard at work on Shiny and, more recently, Shiny Gadgets which
is a version of shiny intended for small interactive work in an RStudio pane or modal. This makes it relatively simple to create interactive UI elements.
The main issue here is that I need to pull data off of an HTML canvas that is not directly controlled by shiny. How do you do this? Well,
you need to use the js Shiny.onInputChange() function to bind your json to Shiny and allow it to be passed back into R as part of Shiny’s
input$... variable. Heres how that is done: note this is the entire interactive portion.
drawMolecule <- function(width=600, height=600) {
ui <- miniPage(
# launch the chemdoodle HTMLWidget
miniContentPanel(chemdoodle_sketcher()),
# add a title bar with a "done" button
gadgetTitleBar("Draw A Molecule", right = miniTitleBarButton("done", "Done", primary = TRUE)),
# use a script to bind the clicking of the done button to
# the recovery of the current molecuel in the Chemdoodle editor (as json)
# binding the JSON to shiny
tags$script('
document.getElementById("done").onclick = function() {
var mol = sketcher.getMolecule();
var jsonmol = new ChemDoodle.io.JSONInterpreter().molTo(mol);
Shiny.onInputChange("moleculedata", jsonmol);};')
)
server <- function(input, output, session) {
# on the server side, this keeps track of the inputs and will return
# the molecule data stored on input$molecule
observeEvent(input$done, { stopApp(input$moleculedata) })
}
runGadget(ui, server,
viewer = dialogViewer("Draw A Molecule",
width = width,
height = height))
}
rJava#
Once the JSON is in R we need to process it as molecule. In the past I had been using the excellent CDK
via the rcdk/rJava packages. To parse the R datastructure we need to loop through the atom and bond elements and add them to an
AtomContainer. We then need to fix the hydrogen on each atom. This was a bit of an effort in getting this to work as there are
some pain points on going from R <–> Java via rJava. But once you get the hang of it, rJava makes it relatively easy to work with Java
interactively, using the $ operator to call methods on R-representations of Java objects. It worked surprisingly well.
I wanted to use CDK 1.5.12 and therefore wanted to update my Mac’s Java. This can cause problems on Yosemite due to a linking issue. You can set the correct link using sudo. Here is how I fixed mine:
# install java 8
sudo ln -f -s $(/usr/libexec/java_home)/jre/lib/server/libjvm.dylib /usr/lib
sudo R CMD javareconf
#from R
install.packages('rJava', type="source")
Once installed, here is a taste of how Java was used:
# define a Java Class for use in R
Bond <- J("org.openscience.cdk.Bond")
# define a new object of a particular class
mol <- .jnew("org.openscience.cdk.AtomContainer")
# loop through your molecule object in R, create some Java bond objects
# and add them to your Java object.
# this looks and behaves a lot like normal R
# you will encounter some type errors though :(
for (bond in moljson$b){
# Note: each index has to be incremented by one becasue lists are 1-indexed
beginindex <- bond$b + 1
endindex <- bond$b + 1
bondorder <- bond$o
newbond <- new(Bond, atoms[[beginindex]], atoms[[endindex]])
if (is.null(bondorder)) bondorder <- 1
if (bondorder == 1) newbond$setOrder(single)
if (bondorder == 2) newbond$setOrder(double)
if (bondorder == 3) newbond$setOrder(triple)
mol$addBond(newbond)
}
Summary#
People often ask about which programming languages are the best and I am a bit agnostic on this point. However, I think its pretty clear that R has a great ecosystem and the data-focused aspect of R in combination with the facilities pushed in large part by the RStudio team are making it easier and easier to make interactive tools for data exploration/data display.