Research
Overview: Functional Genomics and Data-Driven Natural Product Discovery#
My research interests center on the use of genomic tools to discover new natural products. In the Brady Lab we primarily focus on the immense biodiversity of the metagenome and therefore most of my recent work has been related to finding and characterizing gene clusters from environmental DNA (eDNA; in contrast to cultured organisms). So far I have managed to maintain a life as a “damp scientist,” involved in both computational and wet-lab work. The computational work aims to apply the tools of microbial ecology to analyse the biosynthetic capacity of metagenomes while the wetlab work aims to use classic and modern bacterial genetics to speed the recovery and activation of eDNA-derived biosynthetic gene clusters.
Metagenomes Intro#
It is now widely recognized that the vast majority of bacteria in most environments are not readily cultured in the laboratory. The inability to culture these bacteria renders them incompatible with the most heavily relied upon techniques for characterizing bioactive natural products. Although it is still not possible to easily culture most bacteria in the environment, it is possible to extract microbial DNA directly from environmental samples and clone this DNA into model cultured bacteria where, for the first time, it can be functionally characterized. Our group has pioneered the use of culture independent methods to guide the discovery of novel bioactive secondary metabolites from natural microbial communities.
Comparative analysis of Soil Metagenomes (eDNA, metagenomics)#
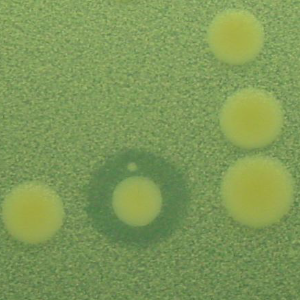
The explosion of DNA sequencing technologies has ignited the field of metagenomics. PCR-based studies using amplicons to assess the biodiversity of microbial communities have indicated that there is a large and largely unexplored bacterial world living in the world all around us, but especially in biodiverse habitats like soil. As bacteria are some of nature’s most prolific and inventive chemists, does this biodiversity mean that there is a corresponding unexplored wealth of small molecule compounds made by organisms that we cannot culture? I address this question using the basic tools of microbial ecology but applying them to conserved domains in the biosynthetic pathways that create interesting small molecules. We aim to use these tools to characterize as many environments as we can.
Homology approaches: Finding Congeners#
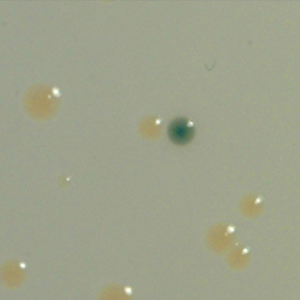
We are developing homology-based approaches for studying gene clusters predicted to encode novel metabolites from within the genomes of uncultured bacteria. With homology screening strategies, clones containing genes known to be involved in the biosynthesis of natural product substructures commonly seen in bioactive compounds are recovered from eDNA libraries, and then each new gene cluster is tested for the ability to encode novel bioactive small molecules. In mysearch for new bioactive small molecules from eDNA libraries, Ihave focused on the recovery of clones from specific small molecule-producing gene clusters.
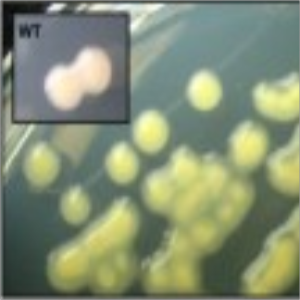
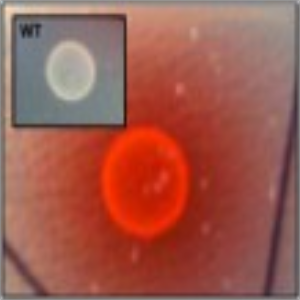
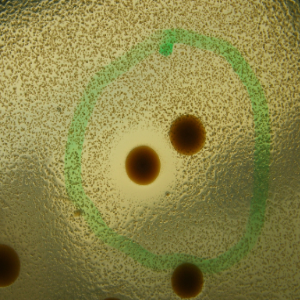
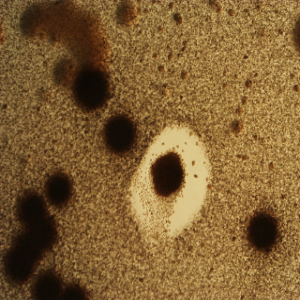
Phenotype Approaches#
The second strategy I am using to identify small molecule producing biosynthetic systems in large eDNA libraries is phenotype (expression-dependent) screening. With this approach, large insert eDNA libraries are examined directly in simple high throughput assays designed to identify clones exhibiting phenotypes traditionally associated with the production of bioactive small molecules (e.g. antibiosis, antifungal activity, cytotoxicity, etc.). In essentially all reported cases, these studies have been carried out in Escherichia coli. I have been workging to expand the phylogenetic diversity of the model bacterial systems available for hosting and screening large eDNA libraries, and develop complementation-based tools to enrich eDNA libraries for small molecule producing gene clusters.