A Simple Peptide Viz Tool in Clojure using CDK
tl;dr don’t write your own chemistry library if a good one exists and CDK is a good one; clojure has good java interop but it still requires you to learn the classes/interfaces in that library; start simple and small
I work in a field at the crossroads of chemistry and biology. The powerful tools of molecular genetics and biochemistry have led to an understanding of how genes and their “gene-products” (proteins) alter and combine basic chemicals to make all of the components that living beings require. In the microbial world, the understanding of these gene-protein-metabolite networks is driving several areas of science where the major conceptual ideas are to either engineer the metabolic systems to maximize the output of a particular metabolite (biofuels, pharmaceutical precursors) or to use the genetic diversity that exists in nature to try to find novel metabolites. Our lab is interested in the latter: we hope to use naturally occurring genetic diversity to find collections of genes making new molecules. While there are lots of tools for visualizing genes, and great tools for plotting particular compounds, there is a neglected middle ground for visualizing all of these things together and I wanted to write a little visualization tool for that purpose.
My initial idea was to make something functional/reactive with javascript and thought I could make something in Clojure that would let me cross compile between the JVM and the browser. I quickly realized, however, that while making chemical graphs is not too difficult, drawing them in a nice way is. So I scaled back my ambition to use an existing Java library for chemistry, the Chemistry Development Kit (CDK) to simply generate peptide structures that can accommodate the many non-natural amino acids you find in Non-Ribosomal Peptides and I ended up creating an small library that can generate pictures of peptides or animated gifs of peptides. It is a far cry from the original intention but it is a minimally viable product of sorts, so I think its a good point to stop and reflect. What follows is a short recap and some thoughts. (all code is in this repo.)
Setup
edit: John May informed me that as of 1.5.10, CDK is available directly from Maven Central. So the EBI repository is no longer necessary
To use CDK from Clojure you need to get the JARs on the classpath. Since February of 2014, CDK is built using Maven and hosted on a repository at the EBI. Big thanks to John May because that conversion makes it as easy to use CDkKas any other JAVA/Clojure library.
In your project.clj file add CDK as a dependency and add the EBI maven repo to your repositories. This will pull in all of CDK. The class dependencies of CDK are tangled making it touch to pull in only what you need.
:dependencies [[org.openscience.cdk/cdk-bundle "1.5.10"]]
:repositories [["ebi-repo"
"http://www.ebi.ac.uk/intact/maven/nexus/content/repositories/ebi-repo"]]
Amino Acid Creation and Peptide Generation
Now my goal was to be able to build peptides to I looked into the ProteinBuilderTool to see if I could generate the protein using a set of sequences. If turns out there is a createProtein function that will build a peptide from canonical amino acids.
BioPolymer protein = ProteinBuilderTool.createProtein("GAGA");
This uses a lookup to find amino acids based on the letter and then will generate a BioPolymer object to which each successive AminoAcid will be added. The amino acids are defined in CML. I don’t have my non-canonical amino acids in CML; I have them in smiles. So my workaround was to create AminoAcids from smiles by 1) parsing the smiles string to create an AtomContainer followed by 2) populate a new AminoAcid instance with the atoms from that AtomContainer while also defining the N- and C- termini (using their position in the smiles string as so):
(def AminoAcids
"map of substrate molecules and their smiles string"
{ ;Canonical amino acids
:ALA "NC(C)C(=O)"
:GLY "NCC(=O)"
:SER "NC(CO)C(=O)"
:THR "NC(C(C)O)C(=O)"
:CYS "NC(CS)C(=O)"
:VAL "NC(C(C)C)C(=O)"
;;; ....
;;; and so on})
(defn- parsesmiles [smiles]
"return an IAtomContainer from smiles input"
(let [builder (SilentChemObjectBuilder/getInstance)
sp (SmilesParser. builder)]
(.parseSmiles sp smiles)))
(defn- ->AminoAcid [mol]
"take an AtomContainer and makes an AminoAcid
the main difference is that AminoAcids have a few extra definitiions that will
be used later that alow finding the N and C termini"
{:pre [(= "N" (.getSymbol (first (.atoms mol))))
(= "C" (last (butlast (.atoms mol))))]}
(let [AA (new AminoAcid)
atoms (seq (.atoms mol))
nterm (.hashCode (first atoms))
cterm (.hashCode (last (butlast atoms)))]
;add atoms
(doseq [a atoms]
(cond
(= nterm (.hashCode a)) (.addNTerminus AA a)
(= cterm (.hashCode a)) (.addCTerminus AA a)
:else (.addAtom AA a)))
;add bonds
(doseq [b (.bonds mol)] (.addBond AA b))
;remove H from C-Terminus
(doto (.getCTerminus AA)
(.setImplicitHydrogenCount (java.lang.Integer. 0)))
AA))
The sharp eyed will notice that I have to remove the H-bond from the C-terminus. This is because, following CDK, I leave out the second oxygen on the C-terminal carbonyl to make adding the peptide bonds easier as you don’t have to remove it later but CDK will add the proton when parsing the smiles string.
Now I have AminoAcids and I want to link them up. I can use the .getNTerminus and .getCTerminus functions but can use clojures sequence abstractions for all of the peptides in my sequence. I can then define the bonds I will have to make and simply make a new AtomContainer to house all of the atoms and bonds, adding the new peptide bonds.
;basic idea of polymer creation
; get the termini, find the bonds to make, and then make them
aaseq (map get-AA aminoacids)
cterms (map #(.getCTerminus %) aaseq)
nterms (map #(.getNTerminus %) aaseq)
bondstomake (partition 2 (interleave (rest nterms) (butlast cterms)))
bonds (map (fn [[a b]] (Bond. a b)) bondstomake)
The Animated Gif Peptide Result
Once the peptide is generated an image needs to be made and I make a clojure port of some basic image making functionality based on CDK’s Standard Generator page. I then used a modification of the gif-clj library to turn a sequence of amino acids into an animated gif of the growing peptide. Heres an example:
(peptideanimation "/Users/zachpowers/Desktop/simplepeptide.gif"
[:PHE :ALA :ASP :GLY] :width 400 :height 800)
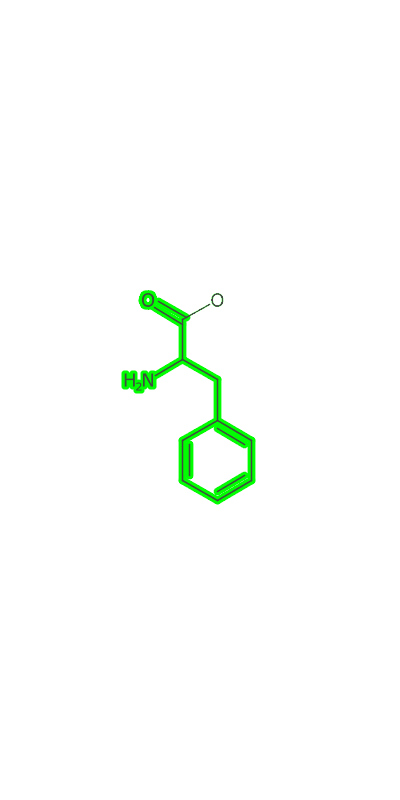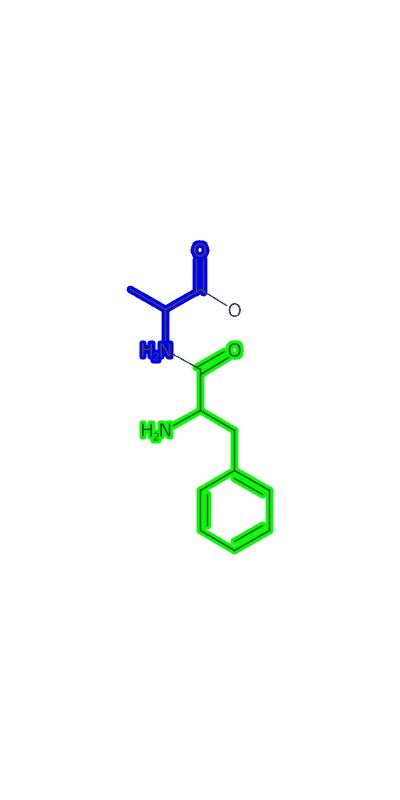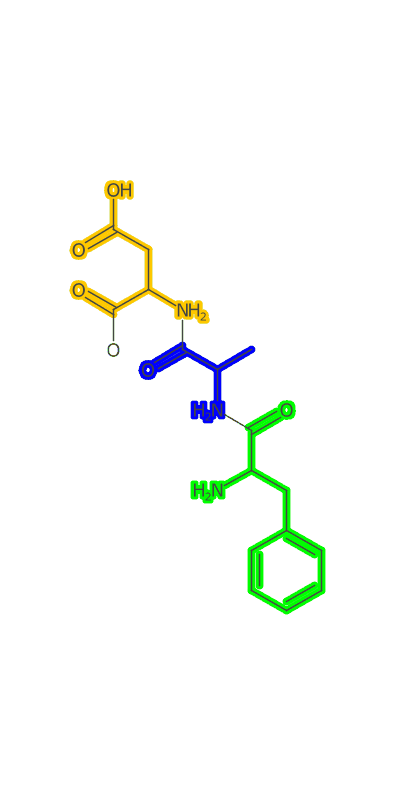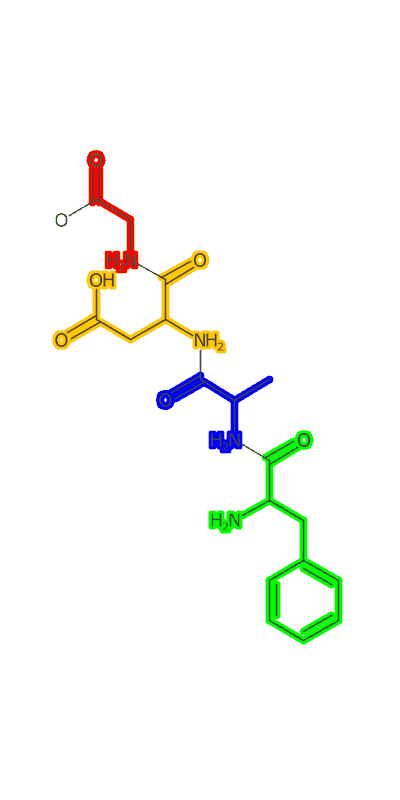
The Take Home
Chemistry libraries take work I started out with big ambitions for making the most awesome biosynthesis visualization library imaginable but quickly ran into some limitations. Most notably: the chemistry knowledge required for making a good chemistry library is nontrivial. Full stop. A library like CDK or RDKit represents a massive amount of work on the part of the developers/scientists. These are mature, fully features libraries built in languages designed for speed and robustness (JAVA/C++). However, they can be a bit of a pain to work with if you want access to the underlying data-structures.
Easy Java Interop for Clojure still requires you to learn the interfaces/classes Most of my time writing this simple app was trying to learn all the various classes and interfaces in the CDK: there are a lot of them. And although Java interop in Clojure is pretty good its not awesome. I’m sure theres better ways to do it but I basically have the Javadocs open in a browser and am poking around looking at the classes to see what they do and how to use them. Usually that requires looking up a dependent class which often has a dependent class … and so on. In IPython, the tab-based introspection helps but really nothing is quite as good as some good documentation/examples. I leaned heavily on John May’s posts to figure out how to do simple things.
Start Simple. Use existing resources My original desire to use Clojure was to make a javascript accessible library that would handle lots of information about genes/proteins/clusters/reactions and have it all be useable in the browser. I now don’t think thats a viable solution. A powerful, mature tool like CDK or RDKit is really needed to generate good looking chemical structures. On the other hand, it feels like you are struggling to work extensively with JAVA - theres a lot of (do ...) and (doseq [...]....) sprinkled about and many layers of classes to get to the values that you want. In the end, the result is very pretty but its tough to learn. I ended up paring back the features and writing a small library that leverages CDK.
So a few thoughts on a possible way forward (if I have some time…..) for making interactive biosynthesis visualizations. First, I think I need to scrap the all-Javascript idea due to the lack of a mature chemistry library on the web. Second, think about ways to interop with a mature library using a small set of functions for which I can write a comprehensive, robust interface. Third, build as much as possible using Clojure data-structures to allow flexibility. If I can manage do that, a functional/reactive component can still be had using server/client interop.
And lastly, its nontrivial to setup a good graphing function. My current code is not very nice from an API perspective and could be made a lot better through some reorganization and some judicious use of macros (see the Incanter charting api for inspiration).